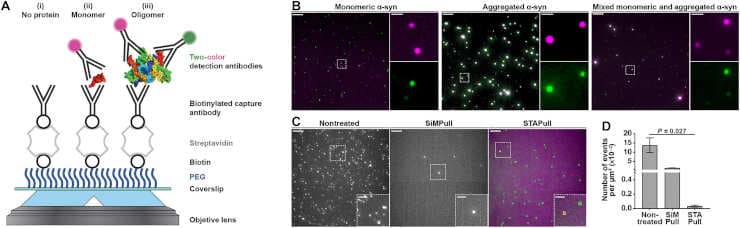
The University of Edinburgh’s Horrocks group has developed a novel solution known as “single-molecule two-color aggregate pull-down,” or STAPull for short, in partnership with UCB Biopharma.
Aggregation — a process in which proteins misfold and group together — is a prominent characteristic of a number of neurological disorders, such as Parkinson’s and Alzheimer’s diseases. These conditions cause tiny, potentially dangerous structures known as oligomers to form, which may be useful markers for an early diagnosis.
They are incredibly small, however, and much rarer than the healthy non-aggregated proteins. This makes it hard to detect and measure them accurately.
STAPull Valuable Protein Aggregate Tool
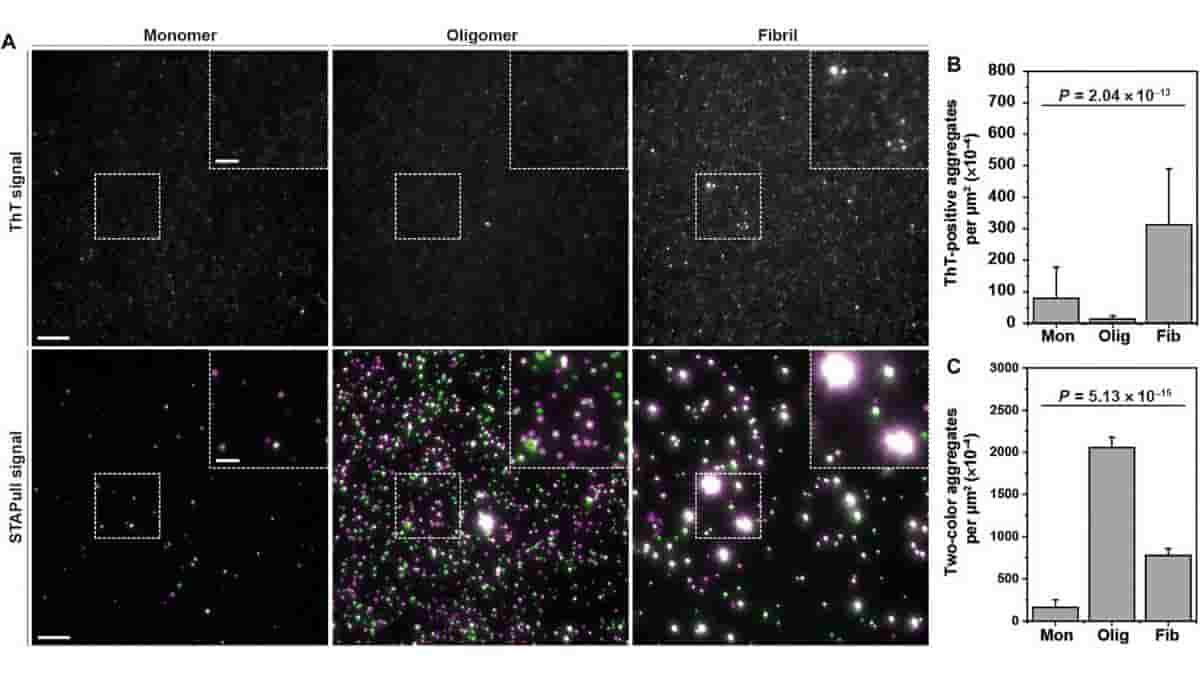
This cutting-edge technique works by examining proteins that have been immobilized (held in place), and labeled with different colors using specific detection antibodies. Researchers can find and measure the proteins that have clumped together while ignoring the ones that are still separate by using sensitive microscopes to look closely at signals where these colors overlap.
Credit: Sci. Adv. 9, eadi7359(2023). DOI:10.1126/sciadv.adi7359
Scientists tested it with the Parkinson’s disease-associated protein alpha-synuclein, and discovered that STAPull could identify these aggregates at physiologically relevant concentrations.
Furthermore, STAPull isn’t limited to a specific type of sample, but can be applied to a wide range of samples, including biofluids from humans. This versatility makes it a valuable tool in the study of protein aggregates associated with various disorders.
Pre-symptomatic Diagnostic Aid
STAPull, by allowing researchers to detect and quantify protein aggregates, opens up new possibilities for identifying biomarkers that can be used to diagnose these debilitating conditions early on, which could be critical in the fight against these diseases.
Currently, patients are diagnosed with neurodegenerative disease based on their symptoms, which appear when the disease is advanced and irreversible cell damage has already occurred. In this work we present an alternative technology, STAPull, that can detect neurodegenerative disease in human biofluids. We are excited to continue developing this technology and explore if it can aid pre-symptomatic diagnosis,”
said lead author, Dr. Rebecca Saleeb, Lady Edith Wolfson Research Fellow, School of Chemistry, University of Edinburgh.
Early detection of neurodegenerative diseases is critical for a wider range of treatment options, improved long-term survival with independence, and improved quality of life.
“Our new technique, STAPull, improved the detection, especially for early stage oligomeric species, which are potentially more harmful but couldn’t be detected with current methods. We are excited to apply this tool to assisting early diagnosis of neurodegenerative diseases in a wide range of samples, including biofluids from humans,”
said lead author, Dr. Ji-Eun Lee. Lee is a Postdoctoral Research Associate in University of Edinburgh’s School of Chemistry.
Abstract
Protein misfolding and aggregation is a characteristic of many neurodegenerative disorders, including Alzheimer’s and Parkinson’s disease. The oligomers generated during aggregation are likely involved in disease pathogenesis and present promising biomarker candidates. However, owing to their small size and low concentration, specific tools to quantify and characterize aggregates in complex biological samples are still lacking. Here, we present single-molecule two-color aggregate pulldown (STAPull), which overcomes this challenge by probing immobilized proteins using orthogonally labeled detection antibodies. By analyzing colocalized signals, we can eliminate monomeric protein and specifically quantify aggregated proteins. Using the aggregation-prone alpha-synuclein protein as a model, we demonstrate that this approach can specifically detect aggregates with a limit of detection of 5 picomolar. Furthermore, we show that STAPull can be used in a range of samples, including human biofluids. STAPull is applicable to protein aggregates from a variety of disorders and will aid in the identification of biomarkers that are crucial in the effort to diagnose these diseases.
Reference:
- Rebecca S. Saleeb et al. Two-color coincidence single-molecule pulldown for the specific detection of disease-associated protein aggregates.Sci. Adv. 9, eadi7359(2023). DOI:10.1126/sciadv.adi7359
Top Image: STAPull detects ThT-negative α-syn oligomers. Credit: Sci. Adv. 9, eadi7359(2023). DOI:10.1126/sciadv.adi7359