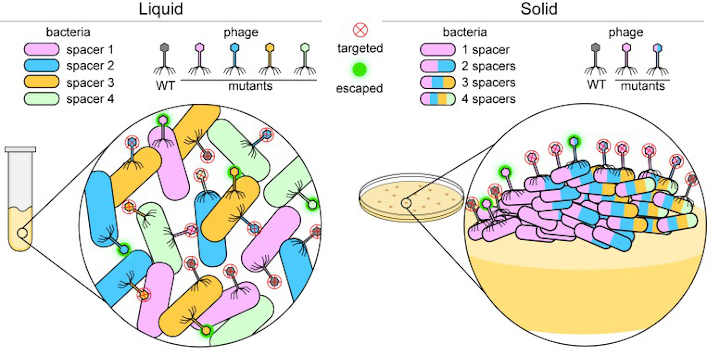
Many bacteria use a system known as CRISPR-Cas to defend themselves against infection by viruses called phages. This system protects the bacterial cell by taking a short length of DNA from the phage and inserting this ‘spacer’ into its own genome.
If the bacterial cell becomes re-infected, the spacer allows the cell to recognize the phage and stop it from replicating by cutting and destroying its DNA. Bacteria with these spacers survive infections and pass their spacers on to their progeny, creating a population that is resistant to the phage.
Phage populations, however, can also adapt and evade bacterial CRISPR-Cas systems. For example, if a phage develops a random mutation in the region targeted by the spacer, it may become undetectable by CRISPR-Cas, leaving it free to replicate and infect other cells (Barrangou et al., 2007; Deveau et al., 2008).
Spacer Sequences
Bacteria can combat these phages by creating multiple spacers that target different regions of the phage genome (van Houte et al., 2016; Figure 1). However, previous studies have shown that bacteria rarely acquire multiple spacers (Heler et al., 2015).
Now, Nora Pyenson and Luciano Marraffini from the Rockefeller University report that the number of spacers each bacterial cell acquires depends on its local environment[1].
Bacteria grown in liquid culture rarely have multiple spacers. Pyenson and Marraffini hypothesized that this is because bacterial cells move more freely when in this environment and are thus able to work together to defend themselves (Figure 1, left). This limits the need for individual cells to have multiple spacers in order to be protected.
To test this theory, Pyenson and Marraffini investigated what happens to infected bacteria that are grown on a solid medium where cell movement is restricted. They found that most cells died, but those that acquired resistance formed separate colonies.
Further experiments showed that compared to bacteria grown in liquid culture, bacterial cells in the resistant colonies had often acquired multiple spacers and were able to fight off phages with other mutations (Figure 1, right).
However, it still remained unclear what drives bacteria to acquire multiple spacers. It was previously shown that acquiring an initial spacer can drive the addition of subsequent spacers through a process called priming (Datsenko et al., 2012; Nussenzweig et al., 2019).
Pyenson and Marraffini found that a disproportionate number of second spacers were taken from DNA regions located close to the initial spacer, which is a hallmark of priming. This suggests that priming enables immobile bacteria to survive mutated phages that have escaped detection by allowing the bacteria to create multiple spacers.
Bacterial Communities
Cells in these resistant colonies arranged themselves into unusually shaped sectors, with the number and type of spacers present varying between cells in each sector. It seems that when bacteria are immobilized, individual cells within the colony must acquire more spacers to resist infection by the mutated phage (Figure 1, right).
The new multi-spacer cells then outgrow the rest of the colony, forming these unusually shaped sectors.
These findings suggest that bacteria cooperate and share the spacer load in a liquid environment. On a solid surface, on the other hand, the bacteria are more independent, and if a cell becomes immune to a mutated phage, it will form a new colony sector with its progeny.
In nature, bacteria are often attached to surfaces. For example, some bacteria live at the bottom of bodies of water, and these bacterial communities may be regularly targeted by phages (Tuson and Weibel, 2013).
Bacteria also gather on biological surfaces when preparing to invade other organisms. For example, the bacteria Streptococcus preferentially bind and form colonies on mucus membrane surfaces in the human body before infecting epithelial cells (Beachey, 1981).
The study by Pyenson and Marraffini reveals how different types of environments may influence the way that bacteria and phages respond to each other and synchronously evolve over time.
CRISPR-Cas systems are exceptionally diverse and use a variety of mechanisms to defend bacteria against infection. Further experiments studying these systems in a variety of environmental contexts will be important to help explain the diverse mechanisms of CRISPR-Cas systems.
[1] Nora C Pyenson, Luciano A Marraffini. Co-evolution within structured bacterial communities results in multiple expansion of CRISPR loci and enhanced immunity. eLife 2020;9:e53078 DOI: 10.7554/eLife.53078
Authors: Michael A Schelling, Dipali G Sashital. © 2020 eLife Sciences Publications Ltd. Republished via Creative Commons Attribution license.
Related Posts:
- A Neurofunctional Schema For Dimensions Of Consciousness
- Organelles Develop From Building Blocks In Random Bursts
- Extravasation Cascade Of Proangiogenic Monocytes To Tumours Detailed
Last Updated on January 7, 2023