According to a new study, reducing the methylation of a critical messenger RNA can increase the migration of macrophages into the brain and alleviate symptoms of Alzheimer’s disease in a mouse model.
The findings, from Rui Zhang of Air Force Medical University in Xian, China, may give a new therapeutic target for Alzheimer’s disease by illuminating a mechanism for the entry of immune cells from the periphery into the brain.
The buildup of proteinaceous, extracellular amyloid-beta plaques in the brain is believed to be one of the causes of Alzheimer’s disease. High amyloid beta levels in mice are associated with neurodegeneration and cognitive symptoms from Alzheimer’s disease in humans, and amyloid beta reduction is a key objective in developing new treatments.
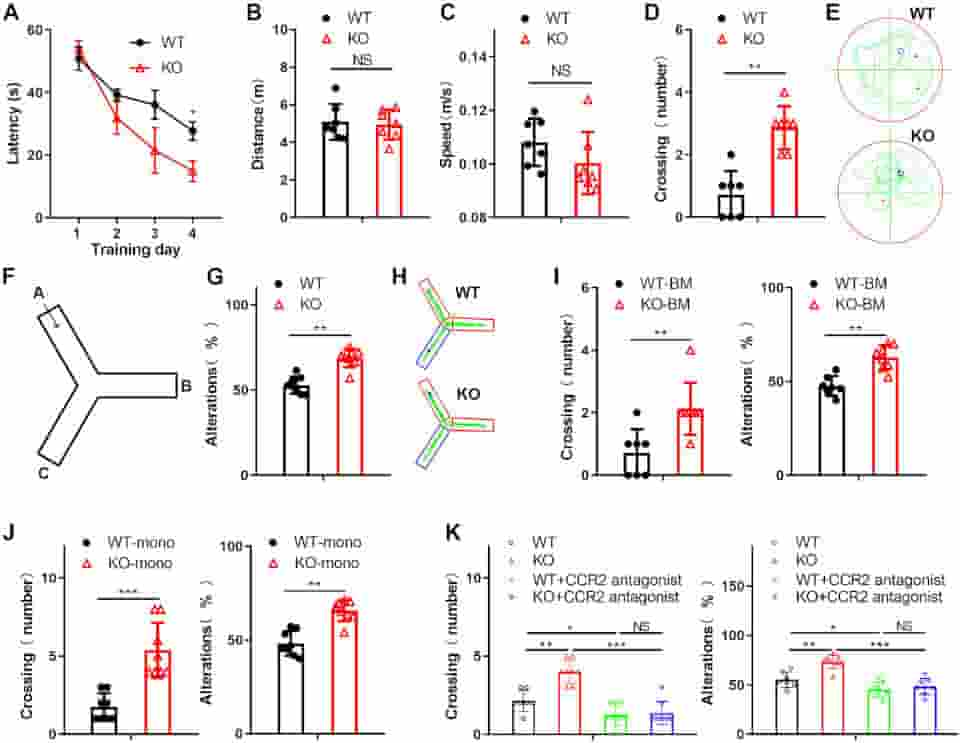
(A). On Day 5, probe trials were performed with the platform absent, and the travel distance (B), swim speed (C), and the number of platform area crossings (D) were assessed. (E) Representative Morris water maze (MWM) swim plots for the WT and KO mice on Day 5.
(F) Experimental design of the Y maze test. (G) The spatial working memory of the WT or KO mice induced by amyloid beta in the Y maze test. Alternations were counted as the percentage of “correct” alternation/total entries. “Correct” alternation means entry into all 3 arms on consecutive choices.
(H) Representative Y maze test plots for the WT and KO mice induced by amyloid beta.
(I) The learning and memory impairment of the amyloid beta-treated WT mice reconstituted with either WT or KO BM was examined by the Morris water maze task and Y maze test
(J) The Morris water maze task and Y maze test examined the spatial learning and memory of mice with WT or KO monocyte transplantation. (K) The Morris water maze task and Y maze test assessed the learning and memory impairment of the amyloid beta-treated WT or KO mice injected with the CCR2 antagonist PF-4136309. All data in the figure are shown as the mean ± SD except for (A). NS means no significant difference.
Credit: PLoS Biol 21(3): e3002017. CC-BY
Myeloid Cell Migration
Myeloid cells are differentiated products of common progenitors derived from bone marrow hematopoietic stem cells. When foreign pathogens attack the body, myeloid cells are rapidly directed into local tissues via different chemokine receptors.
Blood-derived myeloid cells have recently been shown to pass the blood-brain barrier and develop into fully functional macrophages. These myeloid cells are called into action by central nervous system injury and reinforce microglial activities, so they are referred to as blood-derived macrophages
The migration of blood-derived myeloid cells into the brain and their maturation into macrophages, which can devour amyloid-beta with existing microglia, is one potential mechanism for eliminating amyloid beta. That movement is a complex phenomenon governed by numerous interacting players, one of which is the methylation of messenger RNA within myeloid cells.
The enzyme METTL3 is responsible for the most prevalent type of mRNA methylation, known as m6A, so the scientists initially investigated whether METTL3 depletion in myeloid cells affected cognition in the Alzheimer’s disease mice model.
They discovered that it did. Treated mice performed better on several cognitive tests, an effect that could be prevented by preventing myeloid cell migration into the brain.
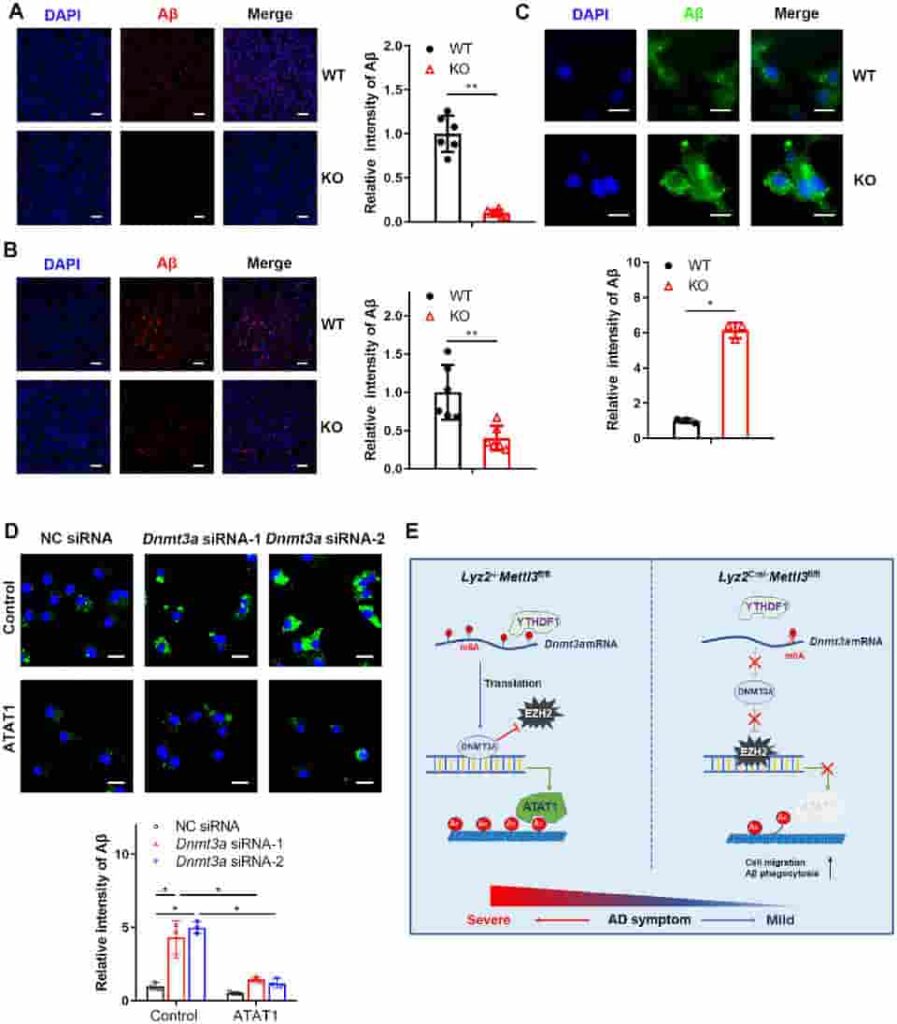
(C) Representative fluorescent micrographs and quantitative analysis of amyloid beta uptake in WT or KO bone marrow-derived macrophages stained with amyloid beta antibody. Scale bars: 10 μm. (D) Representative fluorescence micrographs and quantitative analysis of amyloid beta uptake in the ATAT1-overexpressing THP1 cells transfected with negative control siRNA or Dnmt3a siRNA. Scale bars: 10 μm. (E) Schematic diagram of the role of the m6A writer METTL3 in Alzheimer’s disease.
Credit: PLoS Biol 21(3): e3002017. CC-BY
Lower mRNA Methylation
How does lower mRNA methylation encourage myeloid cell migration? The authors uncovered a complicated mechanism.
They discovered that METTL3 depletion lowered the activity of a crucial m6A reader protein, which detects m6A-modified mRNAs and facilitates their translation into protein, using mRNA expression patterns and other approaches. This resulted in a decrease in another protein, preventing the creation of another protein, ATAT1.
Loss of ATAT1 reduced the attachment of acetyl groups to microtubules, and that reduction, in turn, promoted migration of the myeloid cells across the blood-brain barrier into the brain, followed by maturation into macrophages, increased clearance of amyloid-beta, and improved cognition in mice.
“Our results suggest that m6A modifications are potential targets for the treatment of Alzheimer’s disease,”
the authors concluded.
They noted that much about this pathway in Alzheimer’s disease remains to be explored. Because mRNA methylation broadly impacts a wide range of downstream targets, effective medication development along this route may necessitate further downstream to prevent side effects.
References:
- Yin H, Ju Z, Zheng M, Zhang X, Zuo W, Wang Y, et al. (2023) Loss of the m6A methyltransferase METTL3 in monocyte-derived macrophages ameliorates Alzheimer’s disease pathology in mice. PLoS Biol 21(3): e3002017.
- Kawamoto, H., & Minato, N. (2004). Myeloid cells. The international journal of biochemistry & cell biology, 36(8), 1374–1379.
- Vu, L., Pickering, B., Cheng, Y. et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 23, 1369–1376 (2017)
Last Updated on November 11, 2023