Designing a new generation of drugs against an array of deadly diseases is a costly, arduous and often ineffective task. One of the central challenges is understanding a specific class of proteins on cell surfaces, which are the targets of the majority of pharmaceutical drugs.
Now, using a device known as an X-ray free electron laser (XFEL), Wei Liu and his colleagues at Arizona State University’s Biodesign Institute, along with a team led by Haitao Zhang and Vadim Cherezov of USC have examined one promising drug target in rigorous detail.
The results shed new light on Angiotensin type 2 receptor (ATR2), whose range of functions researchers have only begun to tease out. The group’s findings could speed the development of new compounds addressing cardiovascular conditions, neuropathic pain and tissue growth.
Angiotensin Type 2 Receptor
AT2R belongs to a family of cell receptors known as G-protein coupled receptors (GPCRs), the largest family of cell membrane receptors in the human genome. As Liu explains, GPCRs are such attractive candidates for targeting by new drugs because they provide essential lines of communication between a cell’s external environment and its interior workings.
“Humans have over 800 GPCRs,” Liu says “and every single one of them is important, because they recognize thousands of different entities from outside the cell, including small molecules, peptides, proteins and even photons.”
The light-sensitive GPCR rhodopsin, for example, is critical for vision, olfactory receptors account for the sense of smell, while the caffeine in coffee acts by binding with adenosine receptors, and so forth.
In addition to their vital housekeeping roles, GPCRs provide drug targets for the treatment of a myriad of human illnesses.
“For almost any disease we are struggling with, GPCR proteins could be the potential drug targets: chemokine receptors for HIV, serotonin receptors for Alzheimer’s, dopamine receptors for drug addiction and Parkinson’s,”
Liu says.
Using powerful X-rays from SLAC’s Linac Coherent Light Source (LCLS) and Argonne National Laboratory’s Advanced Photon Source (APS), scientists discovered new structural details of the angiotensin II receptor AT2R that have puzzled researchers for two decades. AT2R is one of two angiotensin II receptors, and its pair, AT1R, has been successfully used as a target for high blood pressure medications.
The information revealed by the study could give drug developers a new path for compounds that combat pain and inflammation or promote tissue regeneration by docking snugly into a pocket in AT2R’s elaborate 3-dimensional shape.
“Unlike its sibling AT1R, the AT2 receptor has not been fully understood. Through this study we uncovered many important details about the AT2 receptor structure and how it binds to chemical compounds, information that will stimulate further studies of the receptor’s function and could enable an exciting next step in drug discovery,”
said Cherezov, professor of chemistry at the University of Southern California and principal investigator of the study.
Two Receptors, Many Mysteries
Both angiotensin II receptors, AT1 and AT2, are key components of a hormone system that helps regulate blood pressure and sodium levels in the blood. They are known as “membrane proteins” because they straddle the plasma membranes of cells, where they receive signals from hormones outside the cell and pass them along to soluble partners inside the cell, such as G proteins or β-arrestins, setting off a cascade of events that spread the signal cell-wide.
Many hypertension medications currently on the market target the AT1 receptor because of its well-understood role in blood pressure regulation; they block AT1 in order to reduce blood pressure. The AT2 receptor, on the other hand, is still an elusive drug target despite multiple studies of its function.
Some, but not all, have shown it counteracts the effects of AT1; others point to its potential for tissue protection and regeneration; and still others suggest it could play a role in blocking pain.
While there are no approved drugs yet that act on AT2 receptors, pharmaceutical companies are aggressively pursuing compounds that will either activate or block them. One such compound, called EMA401, is being tested in patients for the treatment of neuropathic pain.
The latest study addresses two goals: understanding how AT2 differs from AT1, in order to selectively target it ; and exploring why AT2 -which has all the classic features of a GPCR, just like AT1 – fails to activate G-proteins, which spread signals inside cells, or interact with arrestin, turning those signals off.
Ultrafast X-ray Pulses
In the experiments, the researchers looked at two different kinds of samples, which were formed into crystals for examination with X-rays. In one, the AT2 receptor was bound to a selective compound, one that binds only to AT2. These crystals were so small that they could only be studied at SLAC’s X-ray free-electron laser LCLS, where they were streamed across a beam of ultrafast X-ray pulses.
In the other, a non-selective compound was bound to both AT1 and AT2. These samples, which formed larger crystals, were probed at Argonne’s APS synchrotron light source.
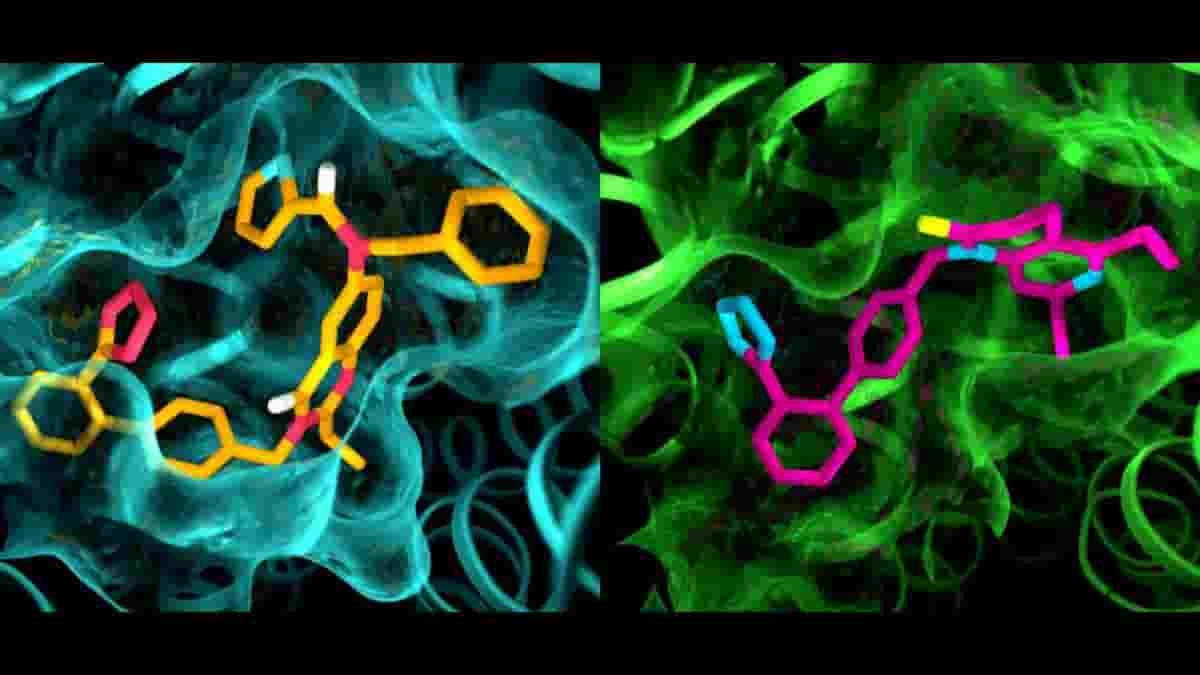
The results of the experiments were surprising in several ways. First, although both compounds were designed to block and deactivate the receptors, they left AT2 in a state that appeared to be active. In addition, although AT1 and AT2 were thought to be very similar, the pockets where the receptors bind to the compounds exhibited marked differences.
“This is something we have never seen with GPCRs,” Cherezov said. “The idea was always that receptors that bind to the same compounds would have very similar pockets, so efforts to develop drugs that act on AT2 started with the same basic structures as drugs that act on AT1. Now we see that we may have to start with entirely different drug-like molecules that are tailored to fit the AT2 receptor, which could set the drug discovery process in a different direction.”
Liu stresses that many important GPCR proteins suitable for drug targets are only able to form very small crystals (under 100 microns in length) in the laboratory. Historically, the ability to grow large protein crystals in the laboratory were a prerequisite for studying their 3d structure at high resolutions, since smaller crystals would be destroyed by synchrotron radiation.
Now however, the study of these molecules has been revolutionized through the use of XFEL technology, which exposes the micron-sized crystals to such short bursts of radiation that they remain intact.
Reference:
- Haitao Zhang, Gye Won Han, Alexander Batyuk, Andrii Ishchenko, Kate L. White, Nilkanth Patel, Anastasiia Sadybekov, Beata Zamlynny, Michael T. Rudd, Kaspar Hollenstein, Alexandra Tolstikova, Thomas A. White, Mark S. Hunter, Uwe Weierstall, Wei Liu, Kerim Babaoglu, Eric L. Moore, Ryan D. Katz, Jennifer M. Shipman, Margarita Garcia-Calvo, Sujata Sharma, Payal Sheth, Stephen M. Soisson, Raymond C. Stevens, Vsevolod Katritch et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature (2017) doi:10.1038/nature22035
Top Image: details in the pocket structures of AT2 (left) and AT1 (right). Credit: Greg Stewart/SLAC National Accelerator Laboratory
Last Updated on December 7, 2023